Tutorial
This page is designed to help you quickly and easily find the information you need. It provides a step-by-step guide on how to use the various features, ensuring an optimal user experience.
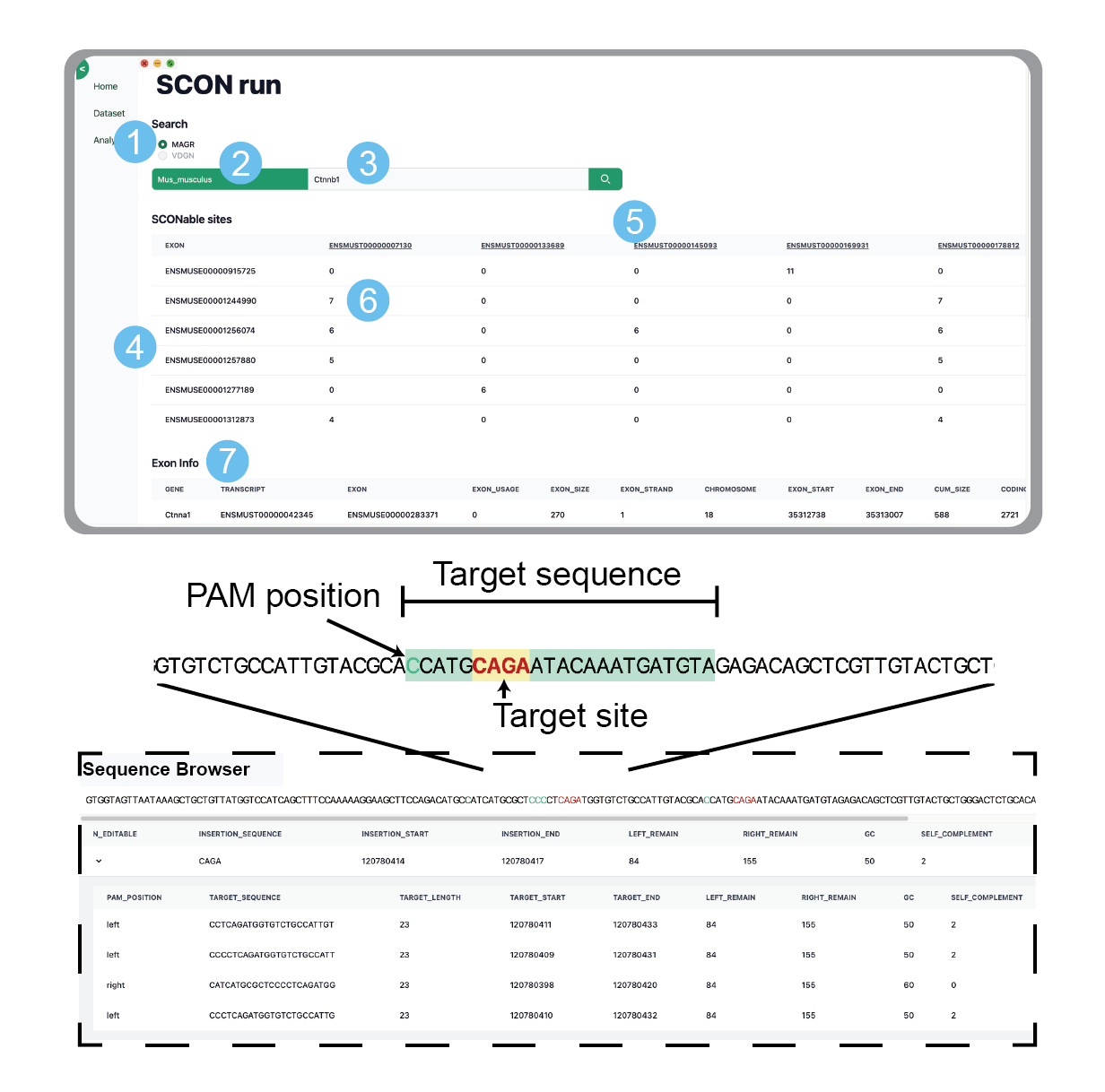
How to use?
STEP 1. Select MAGR or VDGN
This option provides either MAGR((A/C)-A-G-(A/G)) or VDGN((A/G/C)-(A/T/G)-G-(A/T/G/C)) splice junction consensus sequence.
STEP 2. Search for the species you want to insert SCON and click
You can also check and select the desired species from the Dataset menu.
STEP 3. Search for and select the gene you want to insert SCON
When you enter the alphabet of the gene you want, the values related to that alphabet will appear at the bottom of the input window, and you can click on the gene you want.
STEP 4. The exon ID contained in the corresponding gene is located in the leftmost column
Exon ID provides ensembl ID information.
STEP 5. The transcript ID contained in the corresponding gene is located in the top row
The transcript ID provides information about the ensemble ID, and clicking on it takes you to the ensemble site for more information.
STEP 6. Select the cell that matches the exon and transcript you want to insert SCON
The number indicates the number of scon targetable sites.
STEP 7. Contains detailed information about the exons of the selected cell
Clicking on each column will display detailed information about that column.